Catalog # |
Size |
Price |
|
|---|---|---|---|
| 008-89 | 100 µg | $150 |
 )
)
|
Gln-Glu-Ala-Ser-Leu-Phe-Thr-Gly-Pro-Val-Arg
|
| 1203.59 | |
|
| > 97% |
|
| Soluble in water |
|
|
Up to 6 months in lyophilized form at 0-5°C. Long-term storage may form a dimerized peptide. |
|
| Each vial contains 100 µg of NET peptide. |
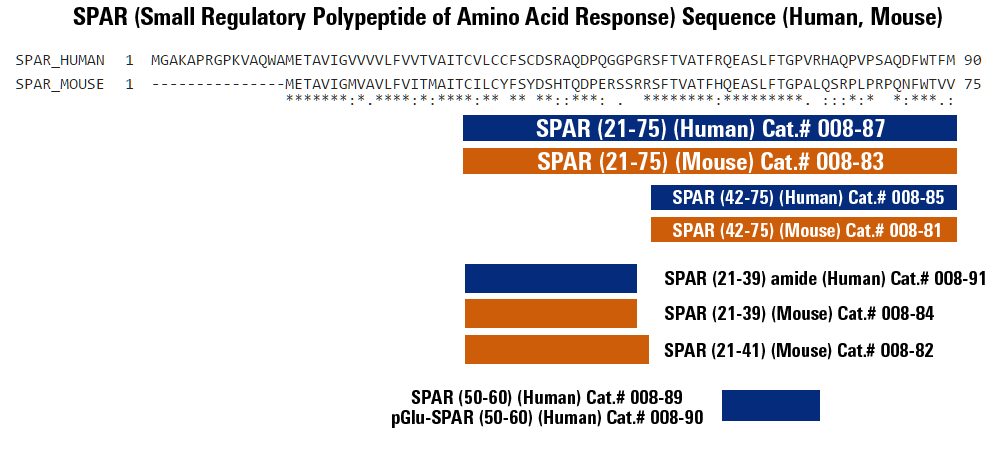
Although prematurely baptized as non-coding, some lncRNAs encode polypeptides with regulatory functions that are implicated in various biological processes. Matsumoto et al. (2017) recently report in Nature that LINC00961 generates SPAR polypeptide that acts via the lysosome to suppress amino-acid-mediated mTORC1 activity, thereby modulating skeletal muscle regenerative response following injury.
Tajbakhsh S. Cell Stem Cell. 2017;20(4):428-430.
Since the vast majority of lncRNAs have been identified by high-throughput computational analysis of RNAseq data, rather than by experimental validation, the question remains as to whether these RNAs are truly “non-coding”. Although one of the criteria for classification of an RNA as long non-coding is that it should have minimal open reading frames (ORFs), and no ORF should encode more than 100 amino acids, short predicted ORFs can be found in many lncRNA molecules. Thus, it is possible that some of these putative lncRNAs may encode for hidden polypeptides. Indeed, there is increasing evidence that suggests that small ORFs within annotated lncRNAs may actually encode for hidden peptides. For example, in 2007 the Kageyama group identified the Drosophila encoded polished rice (pri) lncRNA as encoding 11 or 32 amino acid peptides, [Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 2007; 9:660-5; PMID:17486114; http://dx.doi.org/10.1038/ncb1595] while in 2014 the Schier group identified a hidden polypeptide encoded by the Toddler lncRNA in zebrafish,2Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF. Toddler: An embryonic signal that promotes cell movement via Apelin receptors. [Science 2014; 343:1248636; PMID:24407481; http://dx.doi.org/10.1126/science.1248636] and in 2015 and 2016 the Olson group identified two lncRNA encoded polypeptides regulating the calcium pump SERCA in mammals. [Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance.
Although long non-coding RNAs (lncRNAs) are non-protein-coding transcripts by definition, recent studies have shown that a fraction of putative small open reading frames within lncRNAs are translated. However, the biological significance of these hidden polypeptides is still unclear. Here we identify and functionally characterize a novel polypeptide encoded by the lncRNA LINC00961. This polypeptide is conserved between human and mouse, is localized to the late endosome/lysosome and interacts with the lysosomal v-ATPase to negatively regulate mTORC1 activation. This regulation of mTORC1 is specific to activation of mTORC1 by amino acid stimulation, rather than by growth factors. Hence, we termed this polypeptide 'small regulatory polypeptide of amino acid response' (SPAR). We show that the SPAR-encoding lncRNA is highly expressed in a subset of tissues and use CRISPR/Cas9 engineering to develop a SPAR-polypeptide-specific knockout mouse while maintaining expression of the host lncRNA. We find that the SPAR-encoding lncRNA is downregulated in skeletal muscle upon acute injury, and using this in vivo model we establish that SPAR downregulation enables efficient activation of mTORC1 and promotes muscle regeneration. Our data provide a mechanism by which mTORC1 activation may be finely regulated in a tissue-specific manner in response to injury, and a paradigm by which lncRNAs encoding small polypeptides can modulate general biological pathways and processes to facilitate tissue-specific requirements, consistent with their restricted and highly regulated expression profile.
Matsumoto A, Pasut A, Matsumoto M, et al. Nature. 2017;541(7636):228-232.
No References
| Catalog# | Product | Size | Price | Buy Now |
|---|
Social Network Confirmation