Catalog # |
Size |
Price |
|
|---|---|---|---|
| 029-78 | 100 µg | $393 |
 )
)
|
pGlu-Phe-Gly-Ala-Glu-Cys-Lys-Tyr-Gln-Phe-Gln-Ala-Trp-Gly-Glu-Cys-Asp-Leu-Asn-Thr-Ala-Leu-Lys-Thr-Arg-Thr-Gly-Ser-Leu-Lys-Arg-Ala-Leu-His-Asn-Ala-Glu-Cys-Gln-Lys-Thr-Val-Thr-Ile-Ser-Lys-Pro-Cys-NH2
|
|
Disulfide Bridges: Cys1-Cys3 and Cys2-Cys4 |
| 5354.03 | |
|
| ≥ 95% |
|
| Exhibits correct molecular weight |
|
| Soluble in water |
|
|
Up to 6 months in lyophilized form at 0-5°C.
Long-term storage may form a dimerized peptide For best results, rehydrate just before use. After rehydration, keep solution at +4°C for up to 5 days or freeze at -20°C for up to 3 months. Aliquot before freezing to avoid repeated freeze-thaw cycles. |
|
| White powder |
|
| Each vial contains 100 µg of NET peptide. |
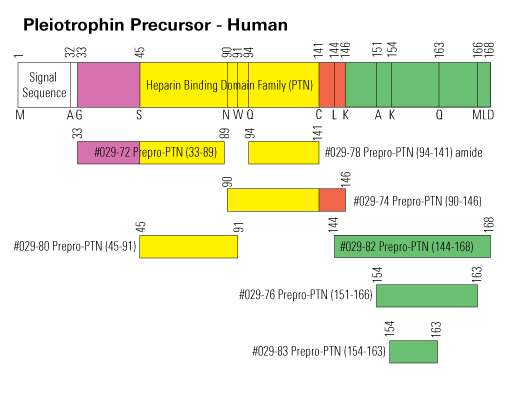

We present a new, quantification-driven proteomic approach to identifying biomarkers. In contrast to the identification-driven approach, limited in scope to peptides that are identified by database searching in the first step, all MS data are considered to choose biomarker candidates. The endopeptidome of cerebrospinal fluid from 40 Alzheimer's disease (AD) patients, 40 subjects with mild cognitive impairment, and 40 controls with subjective cognitive decline was analyzed using multiplex isobaric labeling. Spectral clustering was used to match MS/MS spectra. The top biomarker candidate cluster (215% higher in AD compared to controls, area under ROC curve=0.96) was identified as a fragment of pleiotrophin located near the protein's C-terminus. Analysis of another cohort (n=60 over four clinical groups) verified that the biomarker was increased in AD patients while no change in controls, Parkinson's disease or progressive supranuclear palsy was observed. The identification of the novel biomarker pleiotrophin 151-166 demonstrates that our quantification-driven proteomic approach is a promising method for biomarker discovery, which may be universally applicable in clinical proteomics.
Skillbäck T, Mattsson N, Hansson K, et al. Sci Rep. 2017;7(1):13333.
The lateral ventricle subventricular zone (SVZ) is a frequent and consequential site of pediatric and adult glioma spread, but the cellular and molecular mechanisms mediating this are poorly understood. We demonstrate that neural precursor cell (NPC):glioma cell communication underpins this propensity of glioma to colonize the SVZ through secretion of chemoattractant signals toward which glioma cells home. Biochemical, proteomic, and functional analyses of SVZ NPC-secreted factors revealed the neurite outgrowth-promoting factor pleiotrophin, along with required binding partners SPARC/SPARCL1 and HSP90B, as key mediators of this chemoattractant effect. Pleiotrophin expression is strongly enriched in the SVZ, and pleiotrophin knock down starkly reduced glioma invasion of the SVZ in the murine brain. Pleiotrophin, in complex with the binding partners, activated glioma Rho/ROCK signaling, and ROCK inhibition decreased invasion toward SVZ NPC-secreted factors. These findings demonstrate a pathogenic role for NPC:glioma interactions and potential therapeutic targets to limit glioma invasion.
Qin EY, Cooper DD, Abbott KL, et al. Cell. 2017;170(5):845-859.e19.
Pleiotrophin (PTN) is a heparin-binding growth factor that plays a significant role in tumor growth and angiogenesis. We have previously shown that in order for PTN to induce migration of endothelial cells, binding to both α(ν) β(3) integrin and its receptor protein tyrosine phosphatase beta/zeta (RPTPβ/ζ) is required. In the present study we show that a synthetic peptide corresponding to the last 25 amino acids of the C-terminal region of PTN (PTN(112-136)) inhibited angiogenesis in the in vivo chicken embryo chorioallantoic membrane (CAM) assay and PTN-induced migration and tube formation of human endothelial cells in vitro. PTN(112-136) inhibited binding of PTN to α(ν) β(3) integrin, and as shown by surface plasmon resonance (SPR) measurements, specifically interacted with the specificity loop of the extracellular domain of β(3) . Moreover, it abolished PTN-induced FAK Y397 phosphorylation, similarly to the effect of a neutralizing α(ν) β(3) -choosen antibody. PTN(112-136) did not affect binding of PTN to RPTPβ/ζ in endothelial cells and induced β(3) Y773 phosphorylation and ERK1/2 activation to a similar extent with PTN. This effect was inhibited by down-regulation of RPTPβ/ζ by siRNA or by c-src inhibition, suggesting that PTN(112-136) may interact with RPTPβ/ζ. NMR spectroscopy studies showed that PTN(112-136) was characterized by conformational flexibility and absence of any element of secondary structure at room temperature, although the biologically active peptide segment 123-132 may adopt a defined structure at lower temperature. Collectively, our data suggest that although PTN(112-136) induces some of the signaling pathways triggered by PTN, it inhibits PTN-induced angiogenic activities through inhibition of PTN binding to α(ν) β(3) integrin.
Mikelis C, Lamprou M, Koutsioumpa M, et al. J Cell Biochem. 2011;112(6):1532-43.
BACKGROUND: Pleiotrophin, also known as HARP (Heparin Affin Regulatory Peptide) is a growth factor expressed in various tissues and cell lines. Pleiotrophin participates in multiple biological actions including the induction of cellular proliferation, migration and angiogenesis, and is involved in carcinogenesis. Recently, we identified and characterized several pleiotrophin proteolytic fragments with biological activities similar or opposite to that of pleiotrophin. Here, we investigated the biological actions of P(122-131), a synthetic peptide corresponding to the carboxy terminal region of this growth factor.
RESULTS: Our results show that P(122-131) inhibits in vitro adhesion, anchorage-independent proliferation, and migration of DU145 and LNCaP cells, which express pleiotrophin and its receptor RPTPβ/ζ. In addition, P(122-131) inhibits angiogenesis in vivo, as determined by the chicken embryo CAM assay. Investigation of the transduction mechanisms revealed that P(122-131) reduces the phosphorylation levels of Src, Pten, Fak, and Erk1/2. Finally, P(122-131) not only interacts with RPTPβ/ζ, but also interferes with other pleiotrophin receptors, as demonstrated by choosenknockdown of pleiotrophin or RPTPβ/ζ expression with the RNAi technology.
CONCLUSIONS: In conclusion, our results demonstrate that P(122-131) inhibits biological activities that are related to the induction of a transformed phenotype in PCa cells, by interacting with RPTPβ/ζ and interfering with other pleiotrophin receptors. Cumulatively, these results indicate that P(122-131) may be a potential anticancer agent, and they warrant further study of this peptide.
Diamantopoulou Z, Bermek O, Polykratis A, et al. A Pleiotrophin C-terminus peptide induces anti-cancer effects through RPTPβ/ζ. Mol Cancer. 2010;9:224.
Antibacterial peptides of the innate immune system combat pathogenic microbes, but often have additional roles in promoting inflammation and as growth factors during tissue repair. Midkine (MK) and pleiotrophin (PTN) are the only two members of a family of heparin-binding growth factors. They show restricted expression during embryogenesis and are up-regulated in neoplasia. In addition, MK shows constitutive and inflammation-dependent expression in some non-transformed tissues of the adult. In the present study, we show that both MK and PTN display strong antibacterial activity, present at physiological salt concentrations. Electron microscopy of bacteria and experiments using artificial lipid bilayers suggest that MK and PTN exert their antibacterial action via a membrane disruption mechanism. The predicted structure of PTN, employing the previously solved MK structure as a template, indicates that both molecules consist of two domains, each containing three antiparallel ?-sheets. The antibacterial activity was mapped to the unordered C-terminal tails of both molecules and the last ?-sheets of the N-terminals. Analysis of the highly conserved MK and PTN orthologues from the amphibian Xenopus laevis and the fish Danio rerio suggests that they also harbor antibacterial activity in the corresponding domains. In support of an evolutionary conserved function it was found that the more distant orthologue, insect Miple2 from Drosophila melanogaster, also displays strong antibacterial activity. Taken together, the findings suggest that MK and PTN, in addition to their earlier described activities, may have previously unrealized important roles as innate antibiotics.
Svensson SL, Pasupuleti M, Walse B, et al. J Biol Chem. 2010;285(21):16105-15.
Matrix metalloproteinases (MMPs) exert both pro- and antiangiogenic functions by the release of cytokines or proteolytically generated angiogenic inhibitors from extracellular matrix and basement membrane remodeling. In the Mmp2-/- mouse neovascularization is greatly reduced, but the mechanistic aspects of this remain unclear. Using isotope-coded affinity tag labeling of proteins analyzed by multidimensional liquid chromatography and tandem mass spectrometry we explored proteome differences between Mmp2-/- cells and those rescued by MMP-2 transfection. Proteome signatures that are hallmarks of proteolysis revealed cleavage of many known MMP-2 substrates in the cellular context. Proteomic evidence of MMP-2 processing of novel substrates was found. Insulin-like growth factor binding protein 6, follistatin-like 1, and cystatin C protein cleavage by MMP-2 was biochemically confirmed, and the cleavage sites in heparin affin regulatory peptide (HARP; pleiotrophin) and connective tissue growth factor (CTGF) were sequenced by matrix-assisted laser desorption ionization-time of flight mass spectrometry. MMP-2 processing of HARP and CTGF released vascular endothelial growth factor (VEGF) from angiogenic inhibitory complexes. The cleaved HARP N-terminal domain increased HARP-induced cell proliferation, whereas the HARP C-terminal domain was antagonistic and decreased cell proliferation and migration. Hence the unmasking of cytokines, such as VEGF, by metalloproteinase processing of their binding proteins is a new mechanism in the control of cytokine activation and angiogenesis.
Dean RA, Butler GS, Hamma-kourbali Y, et al. Mol Cell Biol. 2007;27(24):8454-65.
No References
| Catalog# | Product | Size | Price | Buy Now |
|---|
Social Network Confirmation