Catalog # |
Size |
Price |
|
|---|---|---|---|
| 038-64 | 100 µg | $289 |
 )
)
|
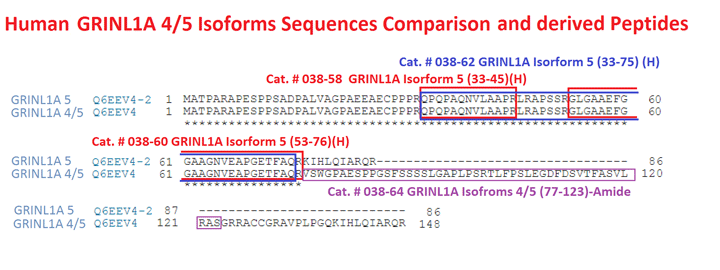
Val-Ser-Trp-Gly-Pro-Ala-Glu-Ser-Pro-Pro-Gly-Ser-Phe-Ser-Ser-Ser-Ser-Leu-Gly-Ala-Pro-Leu-Pro-Ser-Arg-Thr-Leu-Phe-Pro-Ser-Leu-Glu-Gly-Asp-Phe-Asp-Ser-Val-Thr-Phe-Ala-Ser-Val-Leu-Arg-Ala-Ser-NH2
|
| 4798.31 | |
|
| ≥ 95% |
|
| Exhibits correct molecular weight |
|
|
Up to 6 months in lyophilized form at 0-5ºC. For best results, rehydrate just before use. Aliquot before freezing to avoid repeated freeze-thaw cycles. |
|
| White powder |
|
| Each vial contains 100 μg of NET peptide. |
Abstract: Targeting the cell cycle represents a rational approach to mantle cell lymphoma (MCL) therapy, as aberrant expression of cyclin D1 and dysregulation of CDK4 underlie cell cycle progression and proliferation of MCL cells. Although cell cycle cancer therapy was historically ineffective due to a lack of selective and effective drugs, this landscape changed with the advent of selective and potent small-molecule oral CDK4/6 inhibitors. Here, we review the anti-tumor activities and clinical data of selective CDK4/6 inhibitors in MCL. We summarize the known mechanism of action of palbociclib, the most specific CDK4/6 inhibitor to date, and the strategy to leverage this specificity to reprogram MCL for a deeper and more durable clinical response to partner drugs. We also discuss integrative longitudinal functional genomics as a strategy to discover tumor-intrinsic genomic biomarkers and tumor-immune interactions that potentially contribute to the clinical response to palbociclib in combination therapy for MCL. Understanding the genomic basis for targeting CDK4/6 and the mechanisms of action and resistance in MCL may advance personalized therapy for MCL and shed light on drug resistance in other cancers.
Zhao N, Di Liberto M, Elemento O, Chen-Kiang S, Huang X. Disrupting grinl1a association with cereblon accelerates ikzf1/3 degradation and imid killing of mantle cell lymphoma cells by cdk4/6 inhibition. Blood. 2019;134(Supplement_1):299-299.
Abstract: Primary lymphoma of the CNS (PCNSL) is a diffuse large B cell lymphoma confined to the CNS. To elucidate its peculiar organ tropism, we generated recombinant Abs (recAbs) identical to the BCR of 23 PCNSLs from immunocompetent patients. Although none of the recAbs showed self-reactivity upon testing with common autoantigens, they recognized 1547 proteins present on a large-scale protein microarray, indicating polyreactivity. Interestingly, proteins (GRINL1A, centaurin-α, BAIAP2) recognized by the recAbs are physiologically expressed by CNS neurons. Furthermore, 87% (20/23) of the recAbs, including all Abs derived from IGHV4-34 using PCNSL, recognized galectin-3, which was upregulated on microglia/macrophages, astrocytes, and cerebral endothelial cells upon CNS invasion by PCNSL. Thus, PCNSL Ig may recognize CNS proteins as self-Ags. Their interaction may contribute to BCR signaling with sustained NF-κB activation and, ultimately, may foster tumor cell proliferation and survival. These data may also explain, at least in part, the affinity of PCNSL cells for the CNS.
Montesinos-Rongen M, Purschke FG, Brunn A, et al. Primary central nervous system (Cns) lymphoma b cell receptors recognize cns proteins. JI. 2015;195(3):1312-1319.
Abstract: We hypothesized that proteins from the GRINL1A complex transcription unit called Gcom proteins modulate glutamatergic neurotransmission through interaction with the NR1 subunit of the N-methyl D-aspartate (NMDA) receptor. Cotransfection of hemagglutinin-tagged Gcom1 (GRINL1A combined transcript 1) and NR1 cDNAs into HEK293 cells revealed overlapping fluorescent signals in the plasma membrane. Coimmunoprecipitation studies demonstrated reciprocal coimmunoprecipitation from rat brain protein isolates, suggesting an interaction between GRINL1A proteins and the NMDA receptor. Anti-Gcom1 and anti-NR1 antibodies revealed colocalization of postsynaptic immunoreactivity in rat cortical and hippocampal neurons. Finally, anti-Gcom1 antibodies specifically inhibited NMDA excitotoxicity in rat cortical neurons, suggesting a functional interaction of Gcom and NR1 proteins. Our results are consistent with a facilatory role of GRINL1A proteins in glutamatergic signal transduction through interaction with the NMDA receptor.
Roginski RS, Goubaeva F, Mikami M, Fried-Cassorla E, Nair MR, Yang J. GRINL1A colocalizes with N-methyl D-aspartate receptor NR1 subunit and reduces N-methyl D-aspartate toxicity. NeuroReport. 2008;19(17):1721-1726.
Abstract: Our previous proteomics analysis of small proteins expressed in human K562 cells provided the first direct evidence of translation of upstream ORFs in human full-length cDNAs (Oyama, M., Itagaki, C., Hata, H., Suzuki, Y., Izumi, T., Natsume, T., Isobe, T., and Sugano, S. (2004) Analysis of small human proteins reveals the translation of upstream open reading frames of mRNAs. Genome Res. 14, 2048-2052). In the present study, we performed an in-depth proteomics analysis of human K562 and HEK293 cells using a two-dimensional nano-liquid chromatography-tandem mass spectrometry system. The results led to the identification of eight protein-coding regions besides 197 small proteins with a theoretical mass less than 20 kDa that were already annotated coding sequences in the curated mRNA database. In addition to the upstream ORFs in the presumed 5'-untranslated regions of mRNAs, bioinformatics analysis based on accumulated 5'-end cDNA sequence data provided evidence of novel short coding regions that were likely to be translated from the upstream non-AUG start site or from the new short transcript variants generated by utilization of downstream alternative promoters. Protein expression analysis of the GRINL1A gene revealed that translation from the most upstream start site occurred on the minor alternative splicing transcript, whereas this initiation site was not utilized on the major mRNA, resulting in translation of the downstream ORF from the second initiation codon. These findings reveal a novel post-transcriptional system that can augment the human proteome via the alternative use of diverse translation start sites coupled with transcriptional regulation through alternative promoters or splicing, leading to increased complexity of short protein-coding regions defined by the human transcriptome.
Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S. Diversity of translation start sites may define increased complexity of the human short orfeome. Molecular & Cellular Proteomics. 2007;6(6):1000-1006.
No References
| Catalog# | Product | Size | Price | Buy Now |
|---|
Social Network Confirmation